


The clinicalsignificance R
package provides a comprehensive toolkit for analyzing clinical
significance in intervention studies.
Why this package? While statistical significance asks: “Is this effect unlikely due to chance?” Clinical significance asks: “Does this intervention make a meaningful difference for the patient?”
This package empowers researchers and practitioners to move beyond p-values and assess the practical relevance of treatment outcomes.
The package implements the most widely used methods for clinical significance analysis. Each approach answers a specific question:
cs_anchor(): Did the patient
improve by a minimal amount? Evaluates change based on a predefined
Minimal Important Difference (MID).cs_percentage(): Did the patient
improve by a certain percentage? Assesses change relative to the
baseline score.cs_distribution(): Is the change
reliable (beyond measurement error)? Uses statistical distribution
metrics like the Reliable Change Index (RCI).cs_statistical(): Did the patient
return to a functional range? Determines if a patient moved from a
clinical to a functional population.cs_combined(): The “Gold Standard”
(e.g., Jacobson & Truax). Combines reliability and cutoff
criteria for a robust assessment.Install the stable version from CRAN:
install.packages("clinicalsignificance")Or the development version from GitHub:
# install.packages("pak")
pak::pak("benediktclaus/clinicalsignificance")Let’s look at the combined approach (Jacobson &
Truax, 1991). We want to know if patients in the claus_2020
dataset (included in the package) showed a reliable
change AND moved into a functional population
range.
library(clinicalsignificance)
library(ggplot2)
# 1. Perform the analysis
results_combined <- claus_2020 |>
cs_combined(
id = id,
time = time,
outcome = bdi,
pre = 1,
post = 4,
reliability = 0.801,
m_functional = 7.69,
sd_functional = 7.52,
cutoff_type = "c"
)
# 2. Visualize the results
plot(results_combined, show_group = "category")
#> Ignoring unknown labels:
#> • colour : "Group"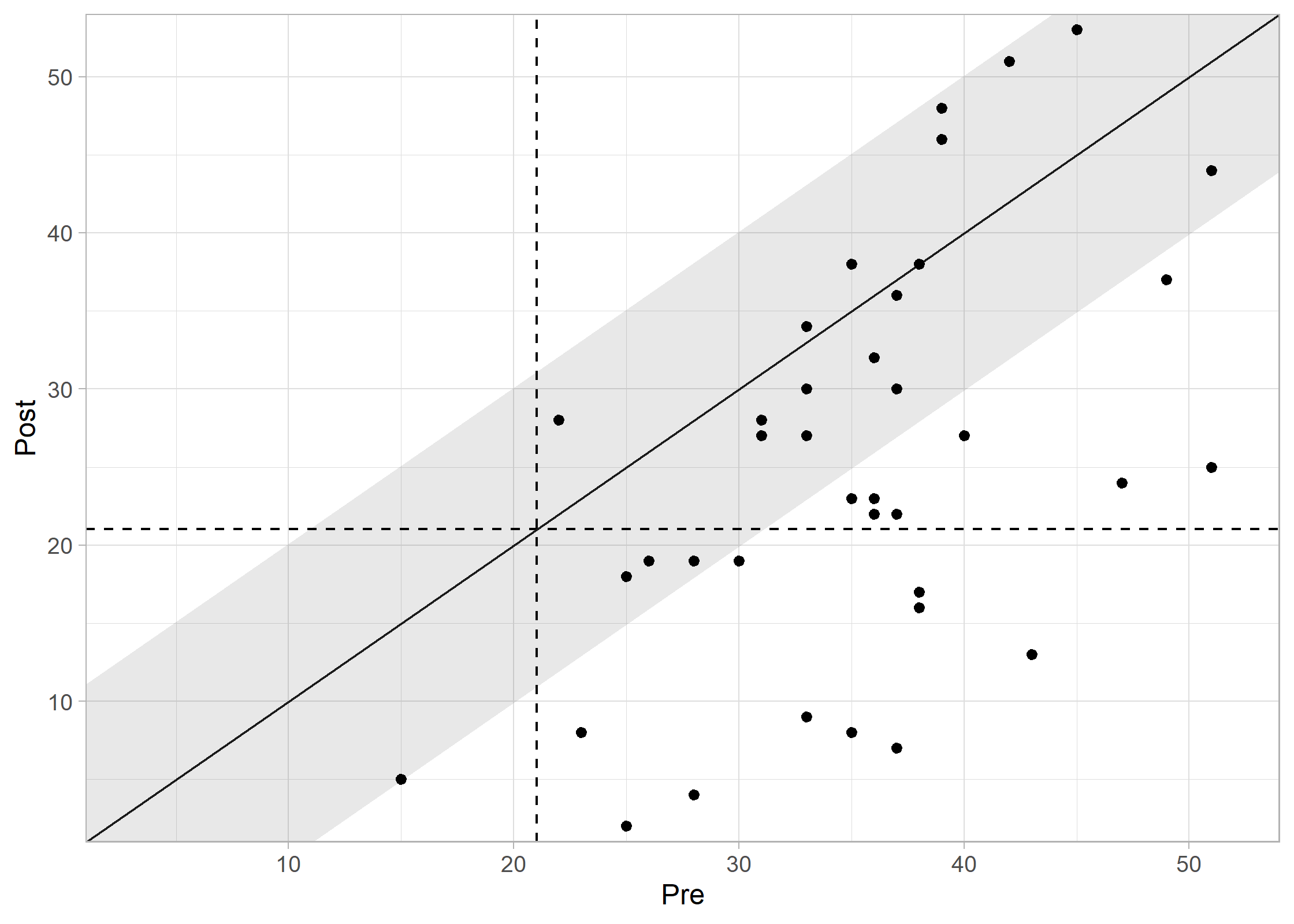
Interpreting the Plot: * Recovered (Green): Reliable improvement + moved to functional range. * Improved (Blue): Reliable improvement, but still in clinical range. * Unchanged (Grey): No reliable change. * Deteriorated (Red): Reliable worsening.
# 3. Get a summary table
summary(results_combined)
#>
#> ---- Clinical Significance Results ----
#>
#> Approach: Distribution-based
#> RCI Method: JT
#> N (original): 43
#> N (used): 40
#> Percent used: 93.02%
#> Outcome: bdi
#> Cutoff Type: c
#> Cutoff: 21.02
#> Outcome: bdi
#> Reliability: 0.801
#>
#> -- Cutoff Descriptives
#>
#> M Clinical | SD Clinical | M Functional | SD Functional
#> -------------------------------------------------------
#> 35.48 | 8.16 | 7.69 | 7.52
#>
#>
#> -- Results
#>
#> Category | N | Percent
#> ---------------------------
#> Recovered | 10 | 25.00%
#> Improved | 8 | 20.00%
#> Unchanged | 22 | 55.00%
#> Deteriorated | 0 | 0.00%
#> Harmed | 0 | 0.00%Please cite both the package and the JSS paper if you use
clinicalsignificance in your research.
Claus, B. B., Wager, J., & Bonnet, U. (2024). clinicalsignificance: Clinical Significance Analyses of Intervention Studies in R. Journal of Statistical Software, 111(1), 1–39. https://doi.org/10.18637/jss.v111.i01
@article{JSS:v111:i01,
author = {Benedikt B. Claus and Julia Wager and Udo Bonnet},
title = {{clinicalsignificance}: Clinical Significance Analyses of Intervention Studies in {R}},
journal = {Journal of Statistical Software},
year = {2024},
volume = {111},
number = {1},
pages = {1--39},
doi = {10.18637/jss.v111.i01},
}
@manual{R-clinicalsignificance,
title = {clinicalsignificance: A Toolbox for Clinical Significance Analyses in Intervention Studies},
author = {Benedikt B. Claus},
year = {2024},
note = {R package version 2.1.0},
doi = {10.32614/CRAN.package.clinicalsignificance},
url = {[https://github.com/benediktclaus/clinicalsignificance/](https://github.com/benediktclaus/clinicalsignificance/)},
}Contributions are welcome! If you encounter bugs or have feature requests: 1. Check the Issue Tracker. 2. Submit a Pull Request.
License: GNU General Public License v3.0
Built with ❤️ for better clinical research.